
- PEPTIDESHAKER DOWNLOAD PDF
- PEPTIDESHAKER DOWNLOAD PORTABLE
- PEPTIDESHAKER DOWNLOAD SOFTWARE
- PEPTIDESHAKER DOWNLOAD DOWNLOAD
PEPTIDESHAKER DOWNLOAD DOWNLOAD
The user can easily select proteins of interest, review the according spectra and download both protein sequences and spectral library.
PEPTIDESHAKER DOWNLOAD SOFTWARE
OkMap 13.7.3 OkMap is a free software for many outdoor activities: Trekking, Off road, Mountain bike, Sailing, Hunting and fishing, Finding mushrooms, Soft air. Price: 75.00, Rating: 5, Downloads: 177 Download. As an additional service, we offer a web service oriented database providing all necessary high-quality and high-resolution data for starting targeted proteomics analyses. PeptideShaker - A viewer for peptide identification results. This system will be designed for high scalability and distributed computing using solutions like the Docker container system among others. In a next step, we are planning to release a web service for protein identification containing both tools. PeptideShaker combines all matches and creates a consensus list of identified proteins providing statistical confidence measures.
PEPTIDESHAKER DOWNLOAD PORTABLE
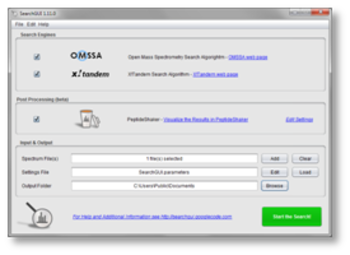
A portable version of this application is available: jPortable. SearchGUI is a managing tool for several search engines to find peptide spectra matches for one or more complex MS2 measurements. Java JRE 10.0.2 / 11 Build 8 Early Access / 8 Build 333. In this work, we present our tools and services, which is the combination of SearchGUI and PeptideShaker. The de.NBI (German network for bioinformatics infrastructure) service center in Dortmund provides several software applications and platforms as services to meet these demands. After certain follow-up analyses, an optional targeted approach is suitable for validating the results. Find, read and cite all the research you need on.
PEPTIDESHAKER DOWNLOAD PDF
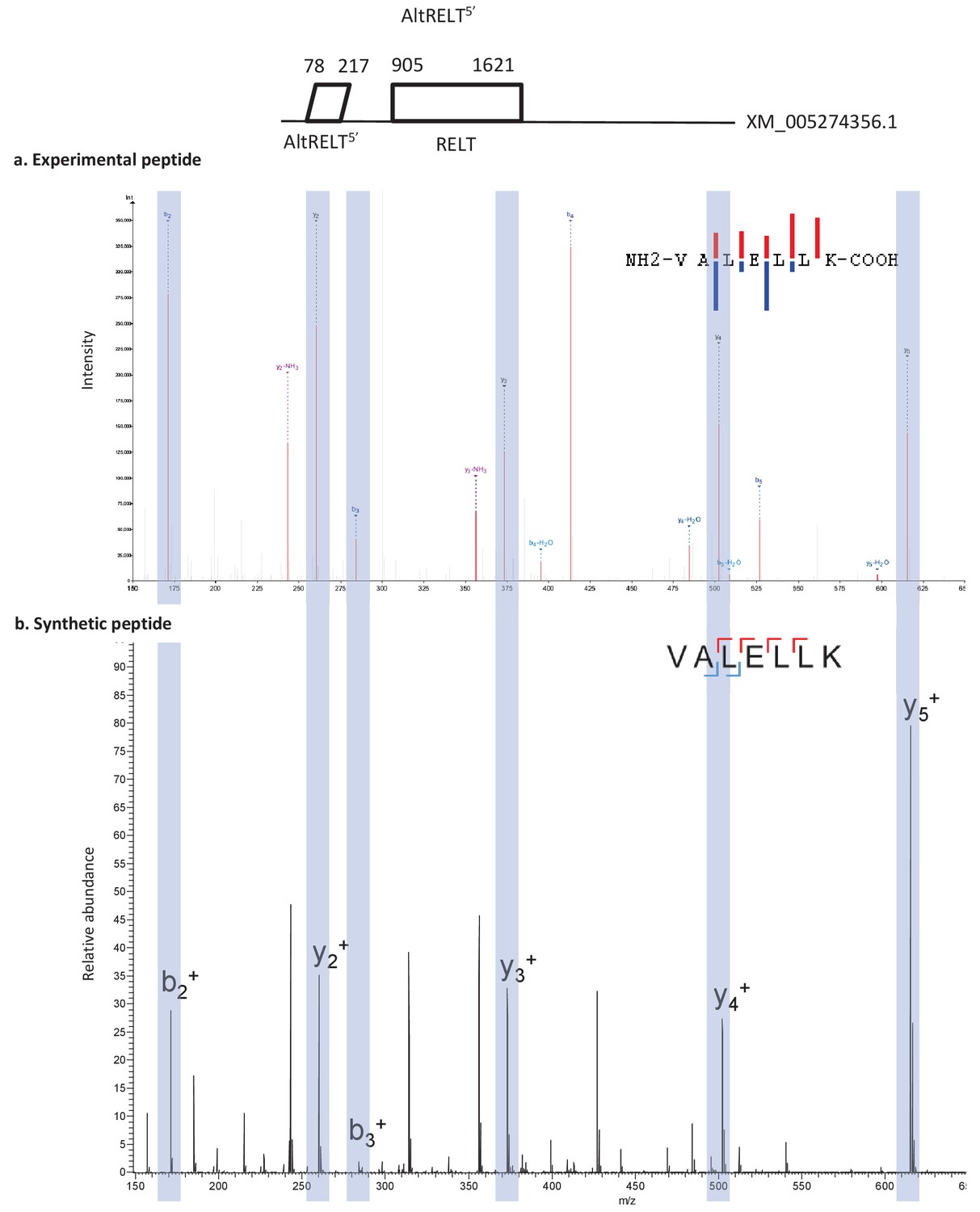
Typically, after capturing with a mass spectrometer, the proteins have to be identified and quantified. PDF Mass spectrometry-based proteomics is a high-throughput technology generating ever-larger amounts of data per project. Part two of the module will be an overview of peptide and protein identification using MS/MS data.Computational proteomics is a constantly growing field to support end users with powerful and reliable tools for performing several computational steps within an analytics workflow for proteomics experiments. Part one of the module will be a brief introduction to the Galaxy framework. Galaxy-P is an extension of Galaxy, a platform developed to enable reproducible data intensive research by providing complex computational software that often requires advanced computer skills via a web-interface.
This two-part module will be an introduction to peptide and protein identification using MS/MS data on the Galaxy-P platform. PeptideShaker is a search engine independent platform for interpretation of proteomics identification results from multiple search engines, currently supporting XTandem, MS-GF+, MS Amanda, OMSSA, MyriMatch, Comet, Tide, Mascot, Andromeda and mzIdentML. It enables you to capture the features offered by various technologies in real time, all with a single click. This however strongly relies on our ability to interpret the massive amounts of data produced by modern mass spectrometers. To start using PeptideShaker, unzip the downloaded file, and double-click the PeptideShaker-X.Y.Z.jar file. Library Sniffer Torrent Download is an extension for Chrome designed to help you identify the web frameworks, javascript libraries and back-end technologies that are currently running on the browsing website. Proteins are then identified by analyzing peptides present in the sample. Mass spectrometry based proteomics studies typically aim at measuring and comparing changes in biological samples. Visualize differential expression and statistical significance using spectrum counting or precursor intensity. Load data from Mascot, Proteome Discoverer, MaxQuant, and more. Search raw data directly with MS Fragger integration and load precursor intensity data from FragPipe. Masses of digested peptides are compared with those predicted from a sequence database to identify peptides. The Industry Standard DDA MS/MS Proteomics Tool. Bottom-up proteomics is a technique for studying proteins that involves digesting proteins to fragment peptides prior to identification using mass spectrometry.


 0 kommentar(er)
0 kommentar(er)
